Data Freezes
freeze-03 (2018-11-01)
Release version: 3.0Version date: 01 November 2018Acronym: freeze-03Release version: 3.0Authors: Michaël Dong, Matthias Hörtenhuber, Abdul Kadir Mukarram, Damir Baranšić, Carsten O. Daub
Highlights
The DANIO-CODE latest data freeze (freeze-03) is now available to the public.
A major extension to our previous data release is remapping to the latest zebrafish genome build (Genome Reference Consortium Zebrafish Build 11, GRCz11). We have also added new data, new assay types (Hi-C) and new pipelines (Hi-C, 3P-seq).
General information
The DANIO-CODE international consortium was established in 2014 in order to generate the most complete functional annotation of the zebrafish genome (Danio Rerio). While zebrafish is widely used in biomedical research, annotation of its reference sequence has fallen behind other important model organisms such as Mus musculus, C. elegans and D. melanogaster following the modENCODE and FANTOM projects.
To bridge this gap, the DANIO-CODE consortium established a Data Coordination Center (DCC) available at https://danio-code.zfin.org. Since 2016, we have collected a large amount of data: 1149 zebrafish sequencing samples, 61 series (55 accessible to public) across 38 developmental stages, 21 assay types and 34 tissue types, and obtained from 31 different research institutes around the world.
The DANIO-CODE DCC aims to provide the largest and most diverse collection of genomic, epigenomic and transcriptomic data for the scientific community to use freely.
Please acknowledge us if you use this data in a publication: danio-code@zfin.org
Data freeze
Data changes with time, as they are replaced, processed and generated by new or updated processing pipelines. Data freezes allow us to capture the current state of the data in our system, including raw and processed data, so that in the future each freeze can be used as a reference point for ongoing analyses. Due to the evolution of the database structure, it is necessary to perform several freezes throughout the DANIO-CODE project to match future major updates; e.g. new data addition or pipeline updates. In short, a data freeze correspond to a referenceable version of our database at a designated stage of the project and that corresponding point of time.
Check the "Additional information" paragraph for the list of datasets (series) that have been included in freeze-03, as well as the main list of changes carried on the data annotation since freeze-02.
freeze-03 contains the following aspects:
- Annotation of experiments and samples (known as metadata)
- Sequencing raw data (e.g. in FASTQ format)
- Data processing pipeline scripts
- Feature tracks to be used in genome browsers e.g. UCSC
- Processed data from our workflow
Please note that for freeze-03, all data uploaded to the DCC are shown, but not all the data is processed or have tracks available. Check the Additional information section for the list of series with processed data available.
Data processing
For each assay type in this freeze, we have developed processing pipelines in order to produce high-quality and standardized ready-for-analysis data. The pipelines are composed of different computational steps, from genome mapping to file formatting and quality control. While pipelines share the same general workflow, they differ in the tools used, the output formats, and the way the signal and peaks are called.
The DANIO-CODE processing workgroup has developed pipelines and processed data for the following assay types:
- RNA-seq pipeline: DANIO-CODE_RNA-Seq_v1.1
- CAGE-seq pipeline: DANIO-CODE_CAGE-seq_pipeline_v1.6
- BS-seq pipeline: DANIO-CODE_BS-seq_v1.6 / DANIO-CODE_WGBS_se_v1.0
- ChIP-seq pipeline: DANIO-CODE_ChIP-seq_cross-correlation_v1.2.1
- ATAC-seq pipeline: DANIO-CODE ATAC-Seq / DNase-Seq Pipeline, v1.0
- MNase-seq pipeline: DANIO-CODE MNase-seq v1.0
- 3P-seq pipeline: DANIO-CODE 3P-seq pipeline v1.0
- Hi-C pipeline: DANIO-CODE_HiC_workflow_v1.0
Note: As requested by our collaborator, the scripts for Hi-C data processing are not publicly available yet. The pipeline code is not yet publicly available but preliminary access is available by contacting Dr Juan M. Vaquerizas at jmv@mpi-muenster.mpg.de.
Mapping to Genome GrCz11
The GRCz11 complete genome assembly from UCSC/Ensembl contains alternate loci scaffolds (ALT_REF_LOCI). We decided to remove these alternative sequences prior to mapping in order to have more uniform coverage distribution.
The edited version of the GrCz11 genome used for our mapping is available at: https://danio-code.zfin.org/freezer/freeze-03/danRer11/danRer11_1_genome/danRer11_1.fa.gz
The script used for removing the alternate loci scaffolds can be found at: https://danio-code.zfin.org/freezer/freeze-03/danRer11/danRer11_1_genome/alt_loci_remove.py
Data availability
The data available to public for freeze-03 includes:
- Sequencing raw data (e.g. in FASTQ format)
- Outputs from our processing pipelines
- UCSC tracks (danRer10 and 11)
Experimental information (metadata) for each dataset is accessible via the data export page of the DCC. To select the data related to freeze-03, select the “freeze-03” Data Version option in the left panel.
Data annotation
With such amounts of data and information to provide, and the differences in how each sequencing sample and dataset are defined, named or structured between institutions, it is necessary to find a way to coordinate all data users working on the same data. Therefore, on the DCC each dataset has to be described following an unique metadata annotation standard, practical, informative and understandable by any DCC data user (https://danio-code.zfin.org/daniocode/help).
When uploading new datasets, the metadata annotation on the DCC is provided by the data provider or a specific annotator, either by importing a formatted csv file (https://danio-code.zfin.org/daniocode/batchUpload/), or via the DCC web interface (https://danio-code.zfin.org/daniocode/addSeries/). If the metadata information is ‘structurally’ complete, the dataset is then uploaded on the DCC with the corresponding metadata annotations associated.
However, successfully uploading the annotations doesn’t mean that the informations given are correct. For each new added dataset (serie) by a data annotator, the data sources are checked, and if there are some issues with the description or the structure that are confirmed, the corrections are applied as soon as possible. The data freeze is not only related to the upload and processing of new datasets; it also includes several metadata mis-annotation corrections carried by the data curator; from one freeze to another, the data can be the same, but described differently.
lease note that the DANIO-CODE DCC is a collaborative effort, and controlling the metadata annotation on the DCC is also based on peer-to-peer reviews. Therefore, if there are any discrepancies in the metadata annotation provided for the datasets, we invite you to contact us, and we will check and solve the issue as soon as possible for the next freeze. Thank you in advance for your support.
Annotations are available on the DCC, on the Data Export page: https://danio-code.zfin.org/dataExport/
Data annotation tutorial: https://danio-code.zfin.org/daniocode/help/
Please note that an user account on the DCC platform is still necessary to view the data annotation on the DCC. If you haven't registered yet, we invite you to sign up and contact the administrator.
Please note that the DANIO-CODE DCC is a collaborative effort, and controlling the metadata annotation on the DCC is also based on peer-to-peer reviews. Therefore, if there are any discrepancies in the metadata annotation provided for the datasets, we invite you to contact us, and we will check and solve the issue as soon as possible for the next freeze. Thank you in advance for your support.
Track hub and Tracks
Track hubs are structured web-accessible genomic datasets that can be visualized online on genome browsers such as UCSC or Ensembl genome browsers. They are very efficient for visualizing large amounts of data without needing to download them.
Tracks for each processed data have been generated and linked to a single track hub aggregating all the signal files available on the DCC. This allow multiple signal files to be displayed in a single custom track, therefore observing multiple types of data or specifically selected ones.
We built custom tracks based on the signal output files from our different processing pipelines. For each sample and/or biosample, a track has been built based on the processed files available, and linked to the track hub.
Tracks for the following assay types are available:
- RNA-seq (UCSC danRer10 & 11)
- 3P-seq (UCSC danRer11)
- CAGE-seq (UCSC danRer10 & 11)
- ChIP-seq (UCSC danRer10 & 11)
- BS-seq (UCSC danRer10)
- ATAC-seq (UCSC danRer10)
- MNase-seq (UCSC danRer10)
- Hi-C (UCSC danRer10 & 11)
Certain tracks required additional conversions of our data. Conversion pipelines are available on Gitlab, in the same repository as the processing pipelines:
- RNA-seq : RNA-seq_signal_conversion_pipeline_v1.0
- CAGE-seq : CAGE-seq_signal_conversion_pipeline_v1.1
The tracks are organised such that a session will display either selected biosample stages consisting of diverse assay types, or selected assay types showing the differences in one biosample stage ( this one will depend on how it will be organised ). Track files associated with the track hub are made available on DANIO-CODE DCC and can be viewed in the UCSC Genome Browser.
Access to freeze-03 files
Link to DANIO-CODE freeze-03 Track hub: https://danio-code.zfin.org/freezer/freeze-03/DANIO-CODE.hub.txt
Tracks for DANIO-CODE freeze-03 are available on the DCC: https://danio-code.zfin.org/freezer/freeze-03
What's next ?
Data since now will be processed on both the Ensembl genome version GrCz10 / UCSC danRer10 and Ensembl genome version GrCz11 / UCSC danRer11 for the mapping steps.
New tracks and track hub version of our processed data are now open to the DCC members for genome visualization. Genome browser users can access and display our generated tracks using this specific format.
We are regularly expanding the range of development stages and biosample types available, by looking into publicly available datasets in the literature, and we also seek your help to complete and enrich the database; if you would like to provide us with more data, we would be glad to be contacted by you.
New pipelines currently developed and datasets will be implemented for freeze-04, as a conclusion for our project. We will also include processed data from ATAC-seq, MNase-seq and BS-seq, remapped to danRer11
If you would like to contribute to our efforts, need help, information or provide any comments or suggestions, we invite you to contact us at the following e-mail address: daniocode@gmail.com.
We would like to thank all the DANIO-CODE consortium partners and colleagues for their support, as well as the collaborators, for providing us with data, suggestions and help for this important stage of the project.
We are grateful for all your contributions. Be sure that we will make our best efforts in carrying this project forward.
With best regards,
Carsten Daub, Ferenc Mueller and Boris Lenhard, on behalf of DANIO-CODE DCC contributors.
DANIO-CODE DCC technical staff:
- Damir Baranašić, pipeline developer (ATAC-seq, BS-seq, ChIP-seq and MNase-seq)
- Michaël Dong, pipeline developer (CAGE-seq), in charge of freeze-02
- Matthias Hörtenhuber, DCC Lead Developer and Administrator
- Abdul Kadir Mukarram, DCC Co-administrator and pipeline developer (RNA-seq)
- Irene Stevens, pipeline developer (3P-seq, ChIP-seq)
- Benjamín Hernández-Rodríguez, pipeline developer (Hi-C)
Additional information
List of series included in freeze-03
| Series name | Series ID/link | Date of raw data upload | Team | Laboratory/Institution, Country | Assay type | Tracks available |
|---|---|---|---|---|---|---|
| ATAC-seq on 24hpf whole embryos | DCD000072SR | May 6th, 2016 | Skarmeta Lab | Centro Andaluz de Biología del Desarrollo, Spain |
|
Yes |
| Genes enriched in pancreatic ductal and beta cells | (private) | May 6th, 2016 | Peers Lab | University of Liège, Belgium |
|
Yes |
| CHIP-seq in early development | DCD000136SR | May 6th, 2016 | Skarmeta Lab | Centro Andaluz de Biología del Desarrollo, Spain |
|
Yes |
| Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. | DCD000137SR | May 6th , 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
No |
| Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. | DCD000138SR | May 6th , 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
No |
| Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. | DCD000139SR | May 6th, 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
Yes |
| Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. | DCD000141SR | May 6th, 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
Yes |
| Upstream ORFs are prevalent translational repressors in vertebrates. | DCD000142SR | May 6th, 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
Yes |
| Eomesodermin targets at sphere stage with RNA-seq | DCD000145SR | May 9th, 2016 | Wardle Lab | King’s College London, UK |
|
Yes |
| Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition | DCD000170SR | May 16th, 2016 | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
|
Yes |
| Zic3 interacts with distant regulatory elements to regulate zebrafish developmental genes | DCD000171SR | May 16th, 2016 | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
|
Yes |
| The Biotagging toolkit for analysis of specific cell populations reveals gene regulatory logic encoded in the nuclear transcriptome (under review) | DCD000177SR | May 18th, 2016 | Sauka-Spengler Lab | University of Oxford, UK |
|
Yes |
| Ribosome Profiling over a Zebrafish Developmental Timecourse | DCD000180SR | May 24th, 2016 | Schier Lab | Harvard University, USA |
|
No |
| Pioneering chromatin for neural crest specification (working title) | (private) | May 30th, 2016 | Sauka-Spengler Lab | University of Oxford, UK |
|
Yes |
| Pou5f3 and Sox2 ChIP-seq | DCD000186SR | May 30th , 2016 | Driever Lab | University of Freiburg, Germany |
|
Yes |
| Zebrafish Globin Locus (DNAse) | DCD000203SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
No |
| Genome-wide maps of binding sites of Nanog-like and Mxtx2 in blastula stage zebrafish embryos | DCD000204SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| A Cdx4-Sall4 regulatory module controls the transition from mesoderm formation to embryonic hematopoiesis | DCD000205SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [zebrafish ChIP-seq] | DCD000207SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| Comprehensive identification of long non-coding RNAs expressed during zebrafish embryogenesis | DCD000225SR | June 9th, 2016 | Schier Lab | Harvard University, USA |
|
Yes |
| DNA methylation reprogramming during early zebrafish embryo development | DCD000227SR | June 11th, 2016 | Liu Lab | Beijing Institute of Genomics, CAS, China |
|
Yes (RNA and BS-seq only) |
| MNase-sequencing of 256-cell and dome embryos to reveal nucleosome organization at promoters during genome activation | DCD000228SR | June 13th, 2016 | Schier Lab | Harvard University, USA |
|
No |
| Comprehensive maps of DNA methylation in mature gametes and at various embryonic stages of cleavage phase zebrafish development. | DCD000229SR | June 16th, 2016 | Cairns Lab | Howard Hughes Medical Institute, USA |
|
Yes |
| Developmental DNA methylation profiling [MethylCap-seq] (zebrafish) | DCD000231SR | July 19th, 2016 | Skarmeta lab | Centro Andaluz de Biologia del Desarrollo |
|
No |
| Active DNA demethylation in zebrafish | DCD000232SR | July 19th, 2016 | Skarmeta lab | Centro Andaluz de Biologia del Desarrollo |
|
No |
| H3K27me3 for 30% epiboly | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Head/trunk CAGE | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| H2A.Z coverage for 30% epiboly | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| CAGE data for developmental time course | DCD000242SR | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Embryonic promoterome | DCD000243SR | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Analysis of open chromatin in early development | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Loss of function of myosin chaperones triggers Hsf1-mediated transcriptional response in skeletal muscle cells | DCD000247SR | Sept. 19th, 2016 | Strähle Lab | Karlsruhe Institute of Technology, Germany |
|
Yes |
| Deep sequencing of small RNA facilitates tissue and sex associated microRNA discovery in zebrafish | DCD000309SR | April 6th, 2017 | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
|
No |
| Comparative analyses of super-enhancers reveal conserved elements in vertebrate genomes | DCD000315SR | April 13th, 2017 | Shkumatava Lab | Institut Curie, France |
|
Yes |
| Genome-wide DNA methylation map of Zebrafish liver (RRBS) | DCD000321SR | April 20th, 2017 | Chatterjee Lab | Dunedin School of Medicine; University of Otago, New Zealand |
|
No |
| Baseline expression from transcriptional profiling of zebrafish developmental stages | DCD000324SR | May 2nd, 2017 | Busch-Nentwich Lab | Wellcome Trust Sanger Institute, UK |
|
Yes |
| RNA-sequencing project for zebrafish embryo and larva development | DCD000328SR | May 9th, 2017 | Yang Lab | Shanghai Chenshan Botanical Garden, China |
|
Yes |
| Eomesodermin and Smad2 targets at high-sphere with ChIP-seq | DCD000332SR | May 19th, 2017 | Wardle Lab | King's College London, UK |
|
Yes |
| RNA-seq data from 4 early developmental stages | DCD000336SR | June 1st, 2017 | Kere Lab | Karolinska Institute, Sweden |
|
Yes |
| Transcriptomic analyses of TCDD treated zebrafish liver | DCD000337SR | June 1st, 2017 | Gong Lab | National University of Singapore |
|
No |
| Transcriptomic analyses of Myc-induced zebrafish liver cancer | DCD000340SR | June 8th, 2017 | Gong Lab | National University of Singapore |
|
No |
| PolyA-seq (3P-Seq) of several stages: Extensive alternative polyadenylation during zebrafish development | DCD000360SR | September 26th, 2017 | Bartel Lab | Whitehead Institute for Biomedical Research, USA |
|
Yes |
| RNA-seq, Ribo-seq and PAL-seq of 3 stages: Poly(A)-tail profiling reveals an embryonic switch in translational control | DCD000370SR | October 9th, 2017 | Bartel Lab | Whitehead Institute for Biomedical Research, USA |
|
No |
| SAPAS Illumina library for 8 developmental stages: "Dynamic landscape of tandem 3’UTRs during zebrafish development" | DCD000371SR | October 23rd, 2017 | Xu Lab | School of Life Sciences, Sun Yat-sen University, China |
|
No |
| RNA-seq, Ribo-seq and PAL-seq of 3 stages: "Conserved Function of lincRNAs in Vertebrate Embryonic Development Despite Rapid Sequence Evolution" | DCD000374SR | October 24th, 2017 | Bartel Lab | Whitehead Institute for Biomedical Research, USA |
|
Yes (ChIP-seq and 3P-seq only) |
| RNA-seq data from heart and mucle, adult stage: "Systematic identification and characterization of cardiac long intergenic noncoding RNAs in zebrafish" | DCD000381SR | December 18th, 2017 | Zhang Lab | University of Maryland, USA |
|
Yes |
| RNA-seq of 12 different adult zebrafish tissues from the Phylofish Database | DCD000382SR | January 8th, 2018 | Bobe lab | Laboratoire de Physiologie et Genomique des Poissons, INRA, Rennes, France |
|
Yes |
| RNA-seq of 6 different tissues: "Annotation of the Zebrafish Genome through an Integrated Transcriptomic and Proteomic Analysis" | DCD000384SR | January 9th, 2018 | Pandey Lab | Johns Hopkins University School of Medicine, USA |
|
Yes |
| RNA-seq of heart: "Telomerase Is Essential for Zebrafish Heart Regeneration" | DCD000388SR | January 9th, 2018 | Gomez Lab | Centro Nacional de Investigaciones Cardiovasculares, Spain |
|
Yes |
| RNA-seq data from 8 different tissues (28dC): "Global identification of the gene networks and cis-regulatory elements of the cold response in zebrafish" | DCD000391SR | January 11th, 2018 | Liangbiao Lab | Shanghai Ocean University, China |
|
Yes |
| RNA-seq of brain for 4 different strains: "Neurotranscriptome profiles of four zebrafish strains" | DCD000392SR | January 12th, 2018 | Wong lab | University of Nebraska Omaha |
|
Yes |
| RNA-seq data from pancreatic cells: "Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes" | DCD000394SR | January 12th, 2018 | Peers Lab | University of Liège, Belgium |
|
Yes |
| RNA-seq of zebrafish brain, liver and skin during perturbation with rotenone at young and old age (control only) | DCD000395SR | January 16th, 2018 | Englert lab | Leibniz Institute for Age Research - Fritz Lipmann Institute |
|
Yes |
| RNAseq from mature ductal cells from nkx6.1:GFP zebrafish lines | DCD000397SR | February 23rd, 2018 | Peers Lab | Peers Lab, Belgium |
|
No |
| RNA-seq data of posterior body: Regulation of posterior body and ectodermal morphogenesis in zebrafish by localized Yap1 and Wwtr1 | DCD000399SR | March 6th, 2018 | Stainier Lab | Max Planck Institute For Heart and Lung Research, Germany |
|
No |
| Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming | DCD000401SR | March 7th, 2018 | Cairns Lab | Howard Hughes Medical Institute, USA |
|
Yes (ChIP-seq only) |
| Ntl, Tbx16 and Mixl1 ChIP-seq data : Genome-wide profiling of Ta, Tbx16 and Mixl1 binding in early zebrafish embryos | DCD000407SR | May 21st, 2018 | Wardle Lab | King's College London, UK |
|
No |
| [NEW] Transcriptome-wide analysis of small RNA expression in early zebrafish development | DCD000416SR | July 9th, 2018 | Wei Lab | Vanderbilt University, USA |
|
No |
| [NEW] ChIP-seq Rad21, RNA-seq comparing WT, Rad21 MO and CTCF MO zebrafish embryos at stages (2.5, 3.3, 4.5, 5.3, 10 hpf) pre and post ZGA and ATAC-seq data: Cohesin facilitates zygotic genome activation in Zebrafish | DCD000426SR | July 16th, 2018 | Horsfield Lab | Department of Pathology, Dunedin School of Medicine, University of Otago |
|
No |
| [NEW] Systemic gain and loss of chromatin architecture throughout zebrafish development | DCD000428SR | July 17th, 2018 | de Wit Lab | Division Of Gene Regulation, Netherlands Cancer Institute |
|
Yes (only Hi-C) |
| [NEW] Expanding the annotation of zebrafish microRNAs based on small RNA sequencing | DCD000429SR | July 20th, 2018 | Postlethwait Lab | Institute of Neuroscience, University of Oregon |
|
No |
| [NEW] ATAC-seq (5 stages) and RNA-seq data (11 stages) : Functional genomic and transcriptomic analysis of amphioxus and the origin of vertebrate genomic traits | DCD000433SR | September 25th, 2018 | Skarmeta Lab | Centro Andaluz de Biologia del Desarrollo |
|
No |
List of data providers / data sources for freeze-03
| Team | Institution | Country |
|---|---|---|
| Lister Lab | University of Western Australia | Australia |
| Peers Lab | University of Liege | Belgium |
| Liangbiao Lab | Shanghai Ocean University | China |
| Liu Lab | Beijing Institute of Genomics, CAS | China |
| Xu Lab | School of Life Sciences, Sun Yat-sen University | China |
| Yang Lab | Shanghai Chenshan Botanical Garden | China |
| Bobe Lab | Laboratoire de Physiologie et Genomique des Poissons, INRA | France |
| Shkumatava Lab | Institut Curie | France |
| Driever Lab | University of Freiburg | Germany |
| Englert Lab | Leibniz Institute for Age Research - Fritz Lipmann Institute | Germany |
| Strahle Lab | Karlsruhe Institute of Technology | Germany |
| Chatterjee Lab | Dunedin School of Medicine; University of Otago | New Zealand |
| Gong Lab | National University of Singapore | Singapore |
| Mathavan Lab | NTU Medical School | Singapore |
| Gomez Lab | Centro Nacional de Investigaciones Cardiovasculares | Spain |
| Skarmeta Lab | Centro Andaluz de Biologia del Desarrollo | Spain |
| Kere Lab | Karolinska Institute | Sweden |
| Busch-Nentwich Lab | Wellcome Trust Sanger Institute | UK |
| Mueller Lab | University of Birmingham | UK |
| Sauka-Spengler Lab | University of Oxford | UK |
| Wardle Lab | King's College London | UK |
| Bartel Lab | Whitehead Institute for Biomedical Research | USA |
| Cairns Lab | Howard Hughes Medical Institute | USA |
| Giraldez Lab | Yale University | USA |
| Pandey Lab | Johns Hopkins University School of Medicine | USA |
| Schier Lab | Harvard University | USA |
| Wong Lab | University of Nebraska Omaha | USA |
| Zhang Lab | University of Maryland | USA |
| Zon Lab | Boston Children's Hospital | USA |
| Wei Lab | Vanderbilt University, Tennessee | USA |
| Horsfield Lab | Department of Pathology, Dunedin School of Medicine, University of Otago | New Zealand |
| de Wit Lab | Division Of Gene Regulation, Netherlands Cancer Institute | Netherlands |
| Postlethwait Lab | Institute of Neuroscience, University of Oregon | USA |
Changelog
- Pipelines
- New pipelines added:
- Hi-C:
- 3P-seq:
- Update of description of track hubs
- Update of description of data curation / metadata annotation
- Adding references
- Documentation for the new pipelines: specific wiki pages / gitlab repositories have been created
- Hi-C pipeline : DANIO-CODE Hi-C
- Data:
- New data series added since freeze-02 (2018-06-01): 5 datasets
- DCD000416SR
- DCD000426SR
- DCD000428SR
- DCD000429SR
- DCD000433SR
- Data series removed since freeze-02 (2018-06-01): 5 datasets
- DCD000232SR
- DCD000323SR
- DCD000208SR
- DCD000209SR
- DCD000206SR
- 3 new assay types
- Hi-C
- small-RNA-seq
- miRNA-seq
- 1 assay type removed
- short-RNA-seq
- Adding track hubs for UCSC Genome Browser (UCSC danRer11):
- RNA-seq (danRer11)
- CAGE-seq (danRer11)
- 3P-seq (danRer11)
- ChIP-seq (danRer11)
- Annotation major changes:
- We noticed that some series were associated to the same publication(s), and thus should be regrouped into the same dataset/series. Subsequently, the following metadata corrections have been applied:
- Series DCD000232SR has been deleted. Related data and annotations have been re-associated to serie DCD000072SR.
- Serie DCD000323SR has been deleted. Related data and annotations have been re-associated to serie DCD000321SR.
- Series DCD000208SR and DCD000209SR have been deleted. Related data and annotations have been re-associated to serie DCD000207SR.
- Serie DCD000206SR has been deleted. Related data and annotations have been re-associated to serie DCD000203SR.
- Missing ATAC-seq data from serie DCD000072SR has been added.
- Some of our data were marked as short RNA-seq. Therefore we added 2 new assay types to be more accurate to the description provided by the publications: micro RNA-seq (miRNA-seq) and small RNA-seq (small-RNA-seq). "short-RNA-seq" has been replaced by "small-RNA-seq". We also corrected some data types for the following series
- DCD000140SR: short-RNA-seq changed to small-RNA-seq
- DCD000309SR: RNA-seq changed to miRNA
- DCD000336SR: RNA-seq changed to small-RNA-seq
- DCD000337SR: RNA-seq changed to small-RNA-seq
- DCD000340SR: RNA-seq changed to small-RNA-seq
- DCD000370SR: RNA-seq changed to miRNA
freeze-02 (2018-06-01)
General information
During the past years, zebrafish has become an increasingly important model organism for research in many areas of biology and medicine. In order to organize and coordinate all the genomics data between different data users, the DANIO-CODE Data Coordination Center (DCC) has been created.
This DANIO-CODE DCC aims to create the largest collection of genomic, epigenomic and transcriptomic data with a thorough annotation of the origins and treatment of samples. We are aiming to become the reference platform for all zebrafish-related data and data metadata, including tracking, storage, processing and distribution to community resources and the scientific community.
Until now, the DANIO-CODE DCC has been used as a platform to gather a large amount of sequencing data, not only from DANIO-CODE members, but also from other public sources.
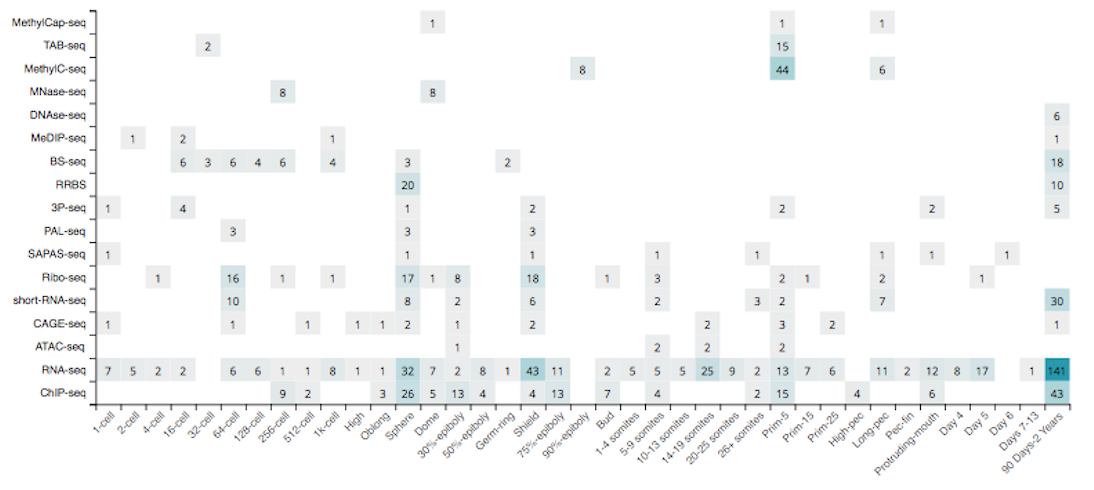
Since the start of the project in 2016, we have collected 980 zebrafish sequencing samples, divided amongst 63 datasets (series) and 17 assay types, covering 38 developmental stages and 34 tissue types, and obtained from 28 different research institutes around the world (May 22th, 2018).
As the volume of data continously increases, we regularly perform data freezes to mark the important checkpoints on the timeline of the project.
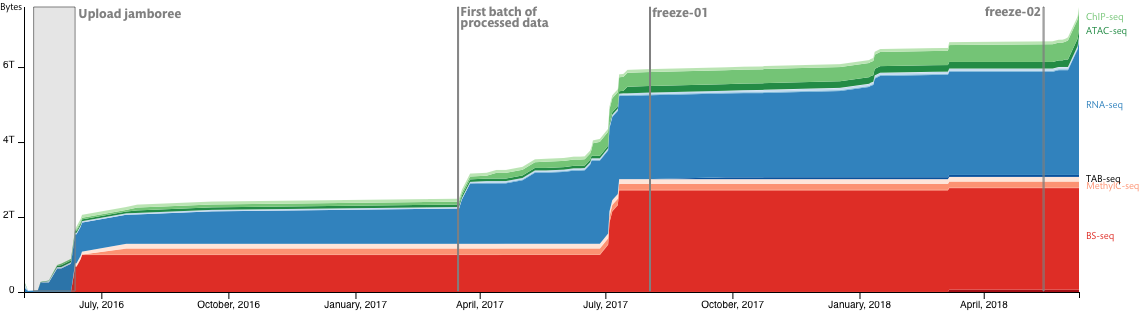
Data freeze
Data changes with time, as they are replaced, processed and generated by new or updated processing pipelines. Data freezes allow us to capture the current state of the data in our system, including raw and processed data, so that in the future each freeze can be used as a reference point for ongoing analyses. Due to the evolution of the database structure, it is necessary to perform several freezes throughout the DANIO-CODE project to match future major updates; e.g. new data addition or pipeline updates. In short, a data freeze correspond to a referenceable version of our database at a designated stage of the project and that corresponding point of time.
After some manual data curation and quality control, we excluded corrupted, incomplete, mis-annotated or relatively low-quality data from the selection. We then pre-processed the raw data with our computational pipelines to obtain some upstream analysis results like mapping outputs against the reference zebrafish genome (Ensembl genome GrCz10). This ready-for-analysis data is made available on the DCC. The data freeze will also be subject of a declaration, through an announcement on the wiki. Each collaborator will then be notified by e-mail of the data freeze release.
See the Additional information section for the list of datasets (series) that have been included in freeze-02.
A freeze contains the following aspects:
- Annotation of experiments and samples (known as metadata)
- Sequencing raw data (e.g. in FASTQ format) [not included in freeze-02]
- Data processing pipeline scripts
- Processed sequencing data [not included in freeze-02]
- Feature tracks to be used e.g. in genome browsers
Please note that for freeze-02, all data uploaded to the DCC are shown, but not all the data is processed or have tracks available. See the Additional information section for the list of series with tracks available.
Data processing
For each assay type in this freeze, we have developed processing pipelines in order to produce high-quality and standardized ready-for-analysis data. Those pipelines are composed of different computational steps, from genome mapping to file formatting and quality control. Those pipelines globally share the same steps, but differ in the tools used, the output formats, and the way the signal and peaks are called.
The DANIO-CODE processing workgroup has developed pipelines for the following assay types (follow the links for more details about each pipeline, their database labels are written after the colon):
- RNA-seq pipeline: DANIO-CODE_RNA-Seq_v1.1
- CAGE-seq pipeline: CAGE_pipeline_v1.6
- BS-seq pipeline: DANIO-CODE_BS-seq_v1.6/DANIO-CODE_WGBS_se_v1.0
- ATAC-seq pipeline: ATAC-Seq / DNase-Seq Pipeline, v1.0
- ChIP-seq pipeline: ChIP-seq_cross-correlation_v1.2.1
- MNase-seq pipeline: DANIO-CODE MNase-seq v1.0
The data have been processed on the DNAnexus cloud-computing platform and on our local servers.
All the pipelines used for processing the data use the same genome GrCz10 / UCSC danRer10 for mapping.
Data availability
The data available to public for freeze-02 includes:
- The curated metadata annotation for each dataset
- The track hub files for UCSC Genome Browser visualization
Data annotation related to freeze-02 are accessible via the data export page of the DCC. To select the data related to freeze-02, select the “freeze-02” Data version option in the left panel.
UCSC track hub / Tracks
Track hubs are structured web-accessible genomic datasets that can be visualized online on genome browsers such as UCSC or Ensembl genome browsers. They are very efficient for visualizing large amounts of data without needing to download them.
Tracks for each processed data have been generated and linked to a single track hub aggregating all the signal files available on the DCC. This allow multiple signal files to be displayed in a single custom track, therefore observing multiple types of data or specifically selected ones.
We built custom tracks based on the signal output files from our different processing pipelines. For each sample and/or biosample, a track has been built based on the processed files available, and linked to the track hub.
Tracks for the following assay types are available:
- RNA-seq
- CAGE-seq
- ChIP-seq
- BS-seq
- ATAC-seq
- MNase-seq
Certain tracks required additional conversions of our data. Conversion pipelines are available on Gitlab, in the same repository as the processing pipelines:
The tracks are organised such that a session will display either selected biosample stages consisting of diverse assay types, or selected assay types showing the differences in one biosample stage (this one will depend on how it will be organised). Track files associated with the track hub are made available on DANIO-CODE DCC and can be viewed in the UCSC Genome Browser.
Access to the DANIO-CODE track hub on UCSC
Go to the UCSC
genome browser, click on the "track hub” button underneath the tracks (or follow this direct link).
There you enter the url of the danio-code track hub: https://danio-code.zfin.org/trackhub/DANIO-CODE.hub.txt, which sends you back to the genome browser with a connection to the DANIO-CODE trackhub.
To make the tracks visible follow these steps:
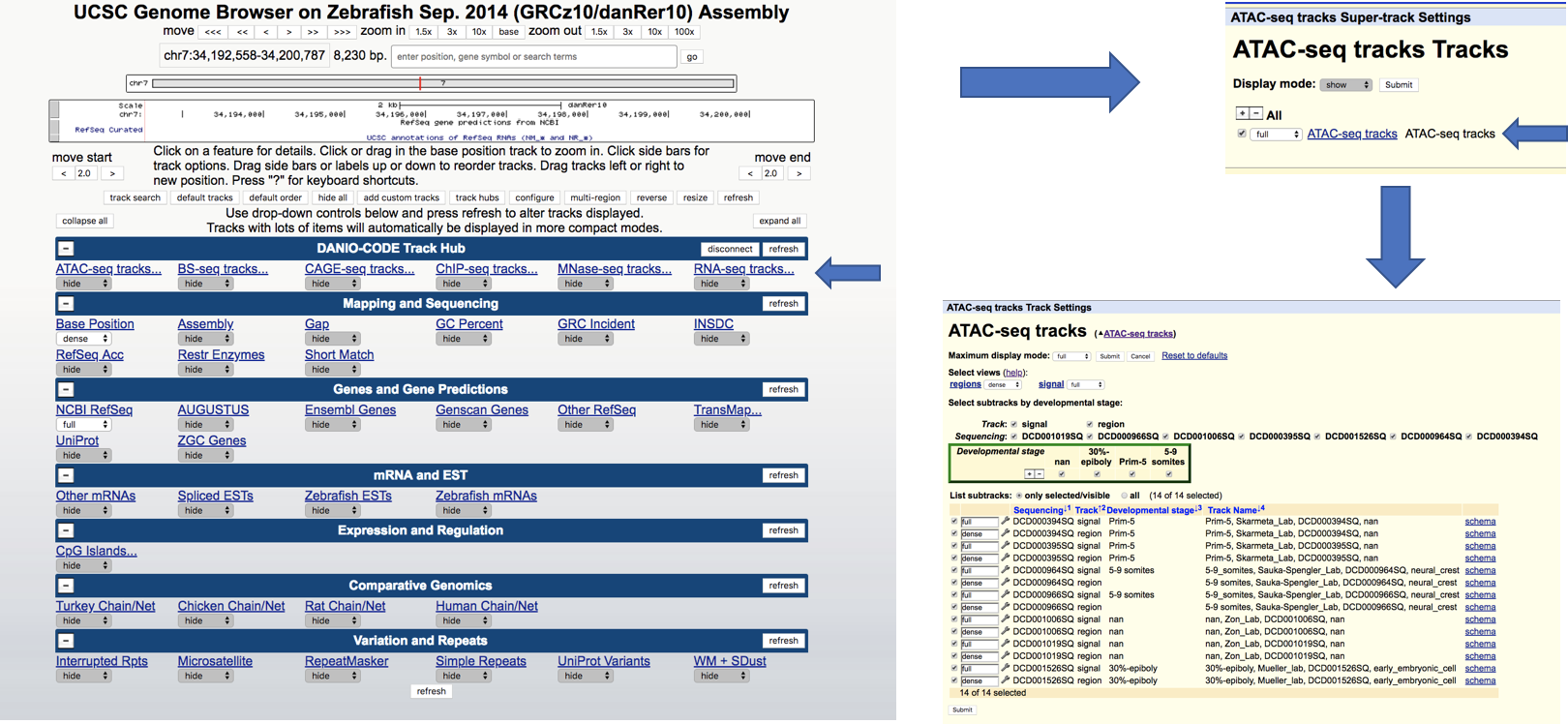
Informations about UCSC track hub: https://genome.ucsc.edu/goldenpath/help/hgTrackHubHelp.html
See the Additional information section for the list of accessible tracks that have been included in freeze-02.
Metadata annotation
Sequencing data by themselves are not descriptive. Each sequencing sample is defined by many characteristics (metadata), such as the type of sequencing data, the developmental stage / hours post-fertilization of the sample, the tissue, the treatments the biosample undergoes, the sequencing instrument used, the date at which it has been sequenced, or the library preparation protocol.
With such amounts of data and information to provide, and the differences in how each sequencing sample and dataset are defined, named or structured between institutions, it is necessary to find a way to coordinate all data users working on the same data. Therefore, on the DCC each dataset has to be described following an unique metadata annotation standard, practical, informative and understandable by any DCC data user (see https://danio-code.zfin.org/daniocode/help).
When uploading new datasets, the metadata annotation on the DCC is provided by the data provider or a specific annotator, either by importing a formatted csv file (https://danio-code.zfin.org/daniocode/batchUpload/), or via the DCC web interface (https://danio-code.zfin.org/daniocode/addSeries/). If the metadata information is ‘structurally’ complete, the dataset is then uploaded on the DCC with the corresponding metadata annotations associated.
However, successfully uploading the annotations doesn’t mean that the informations given are correct. For each new added dataset (serie) by a data annotator, the data sources are checked, and if there are some issues with the description or the structure that are confirmed, the corrections are applied as soon as possible. The data freeze is not only related to the upload and processing of new datasets; it also includes several metadata mis-annotation corrections carried by the data curator; from one freeze to another, the data can be the same, but described differently.
Please note that the DANIO-CODE DCC is a collaborative effort, and controlling the metadata annotation on the DCC is also based on peer-to-peer reviews. Therefore, if there are any discrepancies in the metadata annotation provided for the datasets, we invite you to contact us at the e-mail address indicated below, and we will check and solve the issue as soon as possible for the next freeze. Thank you in advance for your support.
Annotations are available on the DCC, on the data export page: https://danio-code.zfin.org/dataExport/
Data annotation tutorial: https://danio-code.zfin.org/daniocode/help/
Please note that an user account on the DCC platform is still necessary to view the data annotation on the DCC. If you haven't registered yet, we invite you to sign up and contact the administrator.
Update 2018-06-24: A user account on the DCC is not necessary anymore to download data. A user account is only necessary to upload annotations and data.
What is next...
The DANIO-CODE DCC is still a work in progress. You will notice the data available for the freeze is very unequal in terms of quantity and quality from one category to another.
Tracks and trackhub of our processed data are now open to the DCC members for genome visualization. Genome browser users can now access and display our generated tracks using this specific format.
The data available is still being expanded in terms of quantity and quality. Many stages and assay types will be added. We are regularly expanding the range of development stages and biosample types available, by looking into publicly available datasets in the literature, and we also seek your help to complete and enrich the database; if you would like to provide us with more data, we would be glad to be contacted by you.
New pipelines are also being currently developed and will be implemented for freeze-03, making more assay types available.
We also plan to process the data on the new zebrafish Ensembl genome version GrCz11 / UCSC danRer11 for the mapping steps (https://www.ensembl.org/Danio_rerio/Info/Annotation).
If you would like to contribute to this project, need help, information or provide any comments or suggestions, we invite you to contact us at the following e-mail address: daniocode@gmail.com.
We would like to thank all the DANIO-CODE consortium partners and colleagues for their support, as well as the collaborators who provided us with data for this important stage of the project.
We are grateful for all your contributions. Be sure that we will make our best efforts in carrying this project forward.
With best regards,
Carsten Daub, Ferenc Mueller and Boris Lenhard, on behalf of DANIO-CODE DCC contributors.
DANIO-CODE DCC technical staff:
- Damir Baranašić, pipeline developer (ATAC-seq, BS-seq, ChIP-seq and MNase-seq)
- Michaël Dong, pipeline developer (CAGE-seq), in charge of freeze-02
- Matthias Hörtenhuber, DCC Lead Developer and Administrator
- Abdul Kadir Mukarram, DCC Co-administrator and pipeline developer (RNA-seq)
Additional information
List of series included in freeze-02
| Series name | Series ID/link | Date of raw data upload | Team | Laboratory/Institution, Country | Assay type | Tracks available |
|---|---|---|---|---|---|---|
| ATAC-seq on 24hpf whole embryos | DCD000072SR | May 6th, 2016 | Skarmeta Lab | Centro Andaluz de Biología del Desarrollo, Spain |
|
Yes |
| Genes enriched in pancreatic ductal and beta cells | (private) | May 6th, 2016 | Peers Lab | University of Liège, Belgium |
|
Yes |
| CHIP-seq in early development | DCD000136SR | May 6th, 2016 | Skarmeta Lab | Centro Andaluz de Biología del Desarrollo, Spain |
|
Yes |
| Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. | DCD000137SR | May 6th , 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
No |
| Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. | DCD000138SR | May 6th , 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
No |
| Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. | DCD000139SR | May 6th, 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
Yes |
| Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. | DCD000141SR | May 6th, 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
Yes |
| Upstream ORFs are prevalent translational repressors in vertebrates. | DCD000142SR | May 6th, 2016 | Giraldez Lab | Yale School of Medicine, USA |
|
Yes |
| Eomesodermin targets at sphere stage with RNA-seq | DCD000145SR | May 9th, 2016 | Wardle Lab | King’s College London, UK |
|
Yes |
| Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition | DCD000170SR | May 16th, 2016 | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
|
Yes |
| Zic3 interacts with distant regulatory elements to regulate zebrafish developmental genes | DCD000171SR | May 16th, 2016 | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
|
Yes |
| The Biotagging toolkit for analysis of specific cell populations reveals gene regulatory logic encoded in the nuclear transcriptome (under review) | DCD000177SR | May 18th, 2016 | Sauka-Spengler Lab | University of Oxford, UK |
|
Yes |
| Ribosome Profiling over a Zebrafish Developmental Timecourse | DCD000180SR | May 24th, 2016 | Schier Lab | Harvard University, USA |
|
No |
| Pioneering chromatin for neural crest specification (working title) | (private) | May 30th, 2016 | Sauka-Spengler Lab | University of Oxford, UK |
|
Yes |
| Pou5f3 and Sox2 ChIP-seq | DCD000186SR | May 30th , 2016 | Driever Lab | University of Freiburg, Germany |
|
Yes |
| Zebrafish Globin Locus (DNAse) | DCD000203SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
No |
| Genome-wide maps of binding sites of Nanog-like and Mxtx2 in blastula stage zebrafish embryos | DCD000204SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| A Cdx4-Sall4 regulatory module controls the transition from mesoderm formation to embryonic hematopoiesis | DCD000205SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| Zebrafish Globin Locus (ChIP-seq) | DCD000206SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [zebrafish ChIP-seq] | DCD000207SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [zebrafish RNA-Seq] | DCD000208SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [ATAC-Seq] | DCD000209SR | June 2nd, 2016 | Zon Lab | Boston Children Hospital, USA |
|
Yes |
| Comprehensive identification of long non-coding RNAs expressed during zebrafish embryogenesis | DCD000225SR | June 9th, 2016 | Schier Lab | Harvard University, USA |
|
Yes |
| DNA methylation reprogramming during early zebrafish embryo development | DCD000227SR | June 11th, 2016 | Liu Lab | Beijing Institute of Genomics, CAS, China |
|
Yes (RNA and BS-seq only) |
| MNase-sequencing of 256-cell and dome embryos to reveal nucleosome organization at promoters during genome activation | DCD000228SR | June 13th, 2016 | Schier Lab | Harvard University, USA |
|
No |
| Comprehensive maps of DNA methylation in mature gametes and at various embryonic stages of cleavage phase zebrafish development. | DCD000229SR | June 16th, 2016 | Cairns Lab | Howard Hughes Medical Institute, USA |
|
Yes |
| Developmental DNA methylation profiling [MethylCap-seq] (zebrafish) | DCD000231SR | July 19th, 2016 | Skarmeta lab | Centro Andaluz de Biologia del Desarrollo |
|
No |
| Active DNA demethylation in zebrafish | DCD000232SR | July 19th, 2016 | Skarmeta lab | Centro Andaluz de Biologia del Desarrollo |
|
No |
| H3K27me3 for 30% epiboly | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Head/trunk CAGE | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| H2A.Z coverage for 30% epiboly | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| CAGE data for developmental time course | DCD000242SR | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Embryonic promoterome | DCD000243SR | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Analysis of open chromatin in early development | (private) | July 27th, 2016 | Mueller lab | University of Birmingham, UK |
|
Yes |
| Loss of function of myosin chaperones triggers Hsf1-mediated transcriptional response in skeletal muscle cells | DCD000247SR | Sept. 19th, 2016 | Strähle Lab | Karlsruhe Institute of Technology, Germany |
|
Yes |
| Deep sequencing of small RNA facilitates tissue and sex associated microRNA discovery in zebrafish | DCD000309SR | April 6th, 2017 | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
|
No |
| Comparative analyses of super-enhancers reveal conserved elements in vertebrate genomes | DCD000315SR | April 13th, 2017 | Shkumatava Lab | Institut Curie, France |
|
Yes |
| Genome-wide DNA methylation map of Zebrafish liver (RRBS) | DCD000321SR | April 20th, 2017 | Chatterjee Lab | Dunedin School of Medicine; University of Otago, New Zealand |
|
No |
| Genome-wide DNA methylation map of Zebrafish male brain and female brain (RRBS) | DCD000323SR | April 20th, 2017 | Chatterjee Lab | Dunedin School of Medicine; University of Otago, New Zealand |
|
No |
| Baseline expression from transcriptional profiling of zebrafish developmental stages | DCD000324SR | May 2nd, 2017 | Busch-Nentwich Lab | Wellcome Trust Sanger Institute, UK |
|
Yes |
| RNA-sequencing project for zebrafish embryo and larva development | DCD000328SR | May 9th, 2017 | Yang Lab | Shanghai Chenshan Botanical Garden, China |
|
Yes |
| Eomesodermin and Smad2 targets at high-sphere with ChIP-seq | DCD000332SR | May 19th, 2017 | Wardle Lab | King's College London, UK |
|
Yes |
| RNA-seq data from 4 early developmental stages | DCD000336SR | June 1st, 2017 | Kere Lab | Karolinska Institute, Sweden |
|
Yes |
| Transcriptomic analyses of TCDD treated zebrafish liver | DCD000337SR | June 1st, 2017 | Gong Lab | National University of Singapore |
|
Yes |
| Transcriptomic analyses of Myc-induced zebrafish liver cancer | DCD000340SR | June 8th, 2017 | Gong Lab | National University of Singapore |
|
No |
| PolyA-seq (3P-Seq) of several stages: Extensive alternative polyadenylation during zebrafish development | DCD000360SR | September 26th, 2017 | Bartel Lab | Whitehead Institute for Biomedical Research, USA |
|
No |
| RNA-seq, Ribo-seq and PAL-seq of 3 stages: Poly(A)-tail profiling reveals an embryonic switch in translational control | DCD000370SR | October 9th, 2017 | Bartel Lab | Whitehead Institute for Biomedical Research, USA |
|
No |
| SAPAS Illumina library for 8 developmental stages: "Dynamic landscape of tandem 3’UTRs during zebrafish development" | DCD000371SR | October 23rd, 2017 | Xu Lab | School of Life Sciences, Sun Yat-sen University, China |
|
No |
| RNA-seq, Ribo-seq and PAL-seq of 3 stages: "Conserved Function of lincRNAs in Vertebrate Embryonic Development Despite Rapid Sequence Evolution" | DCD000374SR | October 24th, 2017 | Bartel Lab | Whitehead Institute for Biomedical Research, USA |
|
Yes (ChIP-seq only) |
| RNA-seq data from heart and mucle, adult stage: "Systematic identification and characterization of cardiac long intergenic noncoding RNAs in zebrafish" | DCD000381SR | December 18th, 2017 | Zhang Lab | University of Maryland, USA |
|
Yes |
| RNA-seq of 12 different adult zebrafish tissues from the Phylofish Database | DCD000382SR | January 8th, 2018 | Bobe lab | Laboratoire de Physiologie et Genomique des Poissons, INRA, Rennes, France |
|
Yes |
| RNA-seq of 6 different tissues: "Annotation of the Zebrafish Genome through an Integrated Transcriptomic and Proteomic Analysis" | DCD000384SR | January 9th, 2018 | Pandey Lab | Johns Hopkins University School of Medicine, USA |
|
Yes |
| RNA-seq of heart: "Telomerase Is Essential for Zebrafish Heart Regeneration" | DCD000388SR | January 9th, 2018 | Gomez Lab | Centro Nacional de Investigaciones Cardiovasculares, Spain |
|
Yes |
| RNA-seq data from 8 different tissues (28dC): "Global identification of the gene networks and cis-regulatory elements of the cold response in zebrafish" | DCD000391SR | January 11th, 2018 | Liangbiao Lab | Shanghai Ocean University, China |
|
Yes |
| RNA-seq of brain for 4 different strains: "Neurotranscriptome profiles of four zebrafish strains" | DCD000392SR | January 12th, 2018 | Wong lab | University of Nebraska Omaha |
|
Yes |
| RNA-seq data from pancreatic cells: "Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes" | DCD000394SR | January 12th, 2018 | Peers Lab | University of Liège, Belgium |
|
Yes |
| RNA-seq of zebrafish brain, liver and skin during perturbation with rotenone at young and old age (control only) | DCD000395SR | January 16th, 2018 | Englert lab | Leibniz Institute for Age Research - Fritz Lipmann Institute |
|
Yes |
| RNAseq from mature ductal cells from nkx6.1:GFP zebrafish lines | DCD000397SR | February 23rd, 2018 | Peers Lab | Peers Lab, Belgium |
|
No |
| RNA-seq data of posterior body: Regulation of posterior body and ectodermal morphogenesis in zebrafish by localized Yap1 and Wwtr1 | DCD000399SR | March 6th, 2018 | Stainier Lab | Max Planck Institute For Heart and Lung Research, Germany |
|
No |
| Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming | DCD000401SR | March 7th, 2018 | Cairns Lab | Howard Hughes Medical Institute, USA |
|
Yes (ChIP-seq only) |
| Ntl, Tbx16 and Mixl1 ChIP-seq data : Genome-wide profiling of Ta, Tbx16 and Mixl1 binding in early zebrafish embryos | DCD000407SR | May 21st, 2018 | Wardle Lab | King's College London, UK |
|
No |
List of data providers / data sources for freeze-02
| Team | Institution | Country |
|---|---|---|
| Lister Lab | University of Western Australia | Australia |
| Peers Lab | University of Liege | Belgium |
| Liangbiao Lab | Shanghai Ocean University | China |
| Liu Lab | Beijing Institute of Genomics, CAS | China |
| Xu Lab | School of Life Sciences, Sun Yat-sen University | China |
| Yang Lab | Shanghai Chenshan Botanical Garden | China |
| Bobe Lab | Laboratoire de Physiologie et Genomique des Poissons, INRA | France |
| Shkumatava Lab | Institut Curie | France |
| Driever Lab | University of Freiburg | Germany |
| Englert Lab | Leibniz Institute for Age Research - Fritz Lipmann Institute | Germany |
| Strahle Lab | Karlsruhe Institute of Technology | Germany |
| Chatterjee Lab | Dunedin School of Medicine; University of Otago | New Zealand |
| Gong Lab | National University of Singapore | Singapore |
| Mathavan Lab | NTU Medical School | Singapore |
| Gomez Lab | Centro Nacional de Investigaciones Cardiovasculares | Spain |
| Skarmeta Lab | Centro Andaluz de Biologia del Desarrollo | Spain |
| Kere Lab | Karolinska Institute | Sweden |
| Busch-Nentwich Lab | Wellcome Trust Sanger Institute | UK |
| Mueller Lab | University of Birmingham | UK |
| Sauka-Spengler Lab | University of Oxford | UK |
| Wardle Lab | King's College London | UK |
| Bartel Lab | Whitehead Institute for Biomedical Research | USA |
| Cairns Lab | Howard Hughes Medical Institute | USA |
| Giraldez Lab | Yale University | USA |
| Pandey Lab | Johns Hopkins University School of Medicine | USA |
| Schier Lab | Harvard University | USA |
| Wong Lab | University of Nebraska Omaha | USA |
| Zhang Lab | University of Maryland | USA |
| Zon Lab | Boston Children's Hospital | USA |
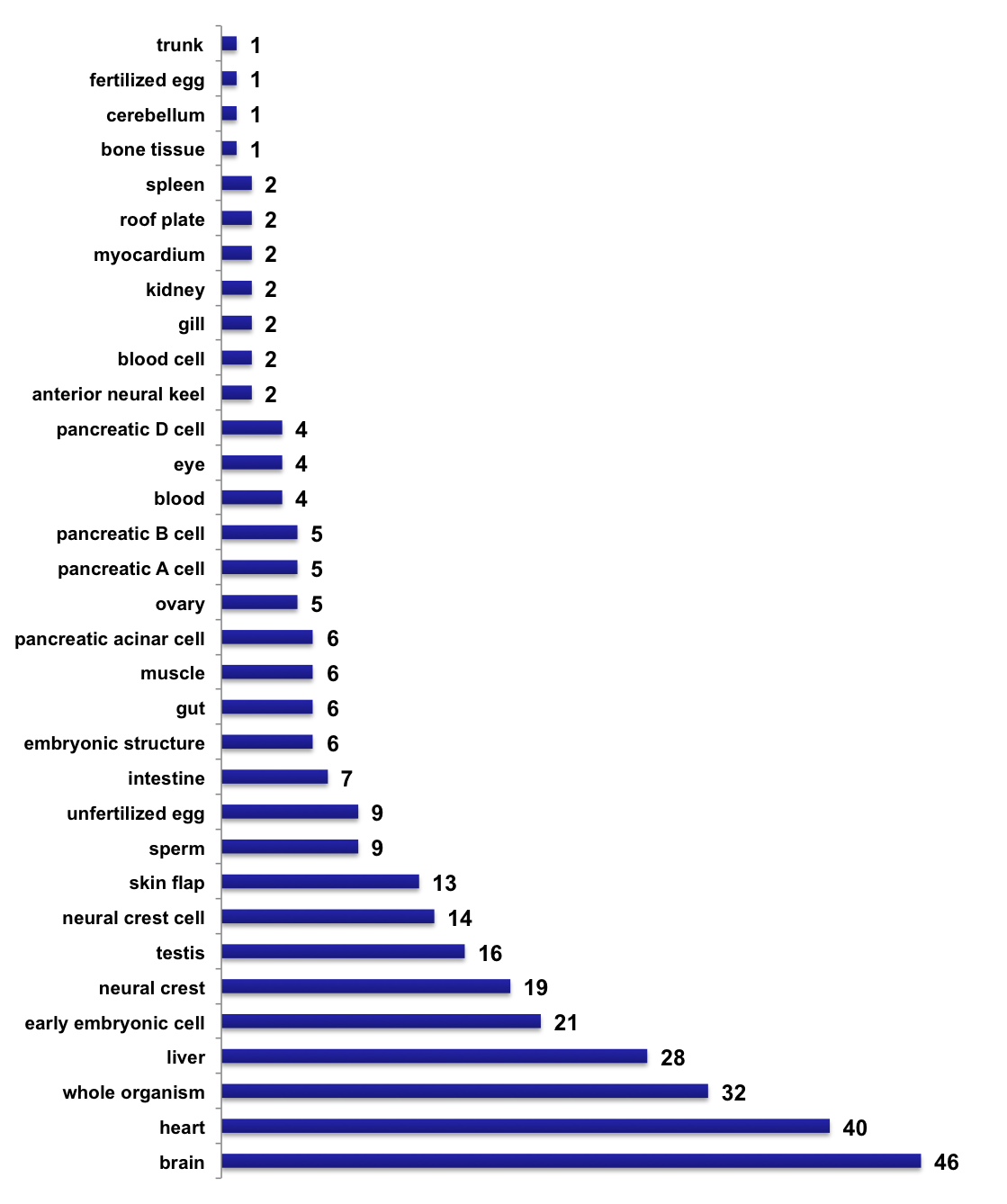
Number of tissue-specific sequencing samples, per tissue type (2018-05-22)
Changelog to freeze-01
- Pipelines
- New pipelines added:
- MNase-seq pipeline : DANIO-CODE MNase-seq v1.0
- Description of track hubs
- Description of data curation / metadata annotation
- Adding references
- Documentation for the new pipelines: specific wiki pages / gitlab repositories have been created
- MNase-seq pipeline : DANIO-CODE MNase-seq v1.0
- Data:
- New data series added since freeze-01 (2017-07-14): 17 datasets
- DCD000340SR
- DCD000360SR
- DCD000370SR
- DCD000371SR
- DCD000374SR
- DCD000381SR
- DCD000382SR
- DCD000384SR
- DCD000388SR
- DCD000391SR
- DCD000392SR
- DCD000394SR
- DCD000395SR
- DCD000397SR
- DCD000399SR
- DCD000401SR
- DCD000407SR
- 3 new assay types
- 3P-seq: Poly(A)-position profiling by sequencing
- PAL-seq: Poly(A)-tail length profiling by sequencing
- SAPAS-seq: Sequencing alternative polyadenylation sites
- Corrections
- Documentation on Gitlab: all annotation curation are now recorded since freeze-01. List of changes can be found on Gitlab
- Adding track hubs for UCSC Genome Browser:
- List tracks/track hubs available
freeze-01 (2017-07-14)
First DANIO-CODE whole-genome data freeze completed
General Information
During the past years, zebrafish has become an increasingly important model organism for research worldwide, for many areas of biology and medicine. In order to organize and coordinate all the genomics data between the different data users, the DANIO-CODE Data Coordination Center (DCC) has been created.
This DANIO-CODE DCC aims to create the largest collection of genomic, epigenomic and transcriptomic data with a thorough annotation of the origins and treatment of samples, looking to become the benchmark platform for all zebrafish-related data and metadata validation, tracking, storage, processing and distribution to community resources and the scientific community.
Until now the DANIO-CODE DCC was used as a platform to gather a large amount of sequencing data, not only from our collaborators, but also from other public sources. Since the start of the project in 2016, we have collected 696 zebrafish sequencing samples, divided amongst 47 datasets (series) and 14 assay types, covering 38 developmental stages, and obtained from 21 different laboratories around the world. As the volume of data increases, it became necessary to perform a data freeze to mark the important checkpoints on the timeline of our project.
Data freeze
Data freezes allow us to capture the current state of the data in our system, including raw and processed data, so that in the future, each freeze can be then used as a reference point for ongoing analyses. Due to the evolution of the database structure, it is necessary to perform several freezes throughout the DANIO-CODE project to match future major updates; e.g. new data addition or pipeline updates.
After some manual data curation and quality control, we selected different sets of data amongst the first ones that have been submitted to the DCC to be part of the freeze; we excluded every corrupted, incomplete, mis-annotated or relatively low-quality data from our selection. We then pre-processed the raw data with our computational pipelines to obtain some upstream analysis results like mapping outputs against the reference zebrafish genome (Ensembl genome GrCz10). This ready-for-analysis data will be made available on the DCC. The data freeze will also be subject of a declaration, through an announcement on the wiki. Each collaborator will then be notified by e-mail of the data freeze release.
The assays for the processed data for freeze-01 range from ChIP-seq, RNA-seq, ATAC-seq, RNA seq, and BS-seq. For these assays, sequences have been processed and have been made part of this data freeze.
See the Additional information section for the list of datasets (series) that have been included in freeze-01.
Data processing
For each assay type in this freeze, we have developed processing pipelines in order to produce high-quality and standardized upstream ready-for-analysis data. Those pipelines are composed of different computational steps, from genome mapping to file formatting and quality control. Those pipelines globally share the same steps, but differ in the tools used, the output formats, and the way the signal and peaks are called.
Until now, the DANIO-CODE processing workgroup has developed pipelines for the following assay types (follow the links for more details about each pipeline, their database labels are written after the colon):
- RNA-seq pipeline: DANIO-CODE_RNA-Seq_v1.1
- CAGE-seq pipeline: CAGE_pipeline_v1.6
- BS-seq pipeline: DANIO-CODE_BS-seq_v1.6 /DANIO-CODE_WGBS_se_v1.0
- ATAC-seq pipeline: DANIO-CODE ATAC-Seq / DNase-Seq Pipeline,v1.0
- ChIP-seq pipeline: DANIO-CODE_ChIP-seq_cross-correlation_v1.2.1
The data have been processed on the DNAnexus cloud-computing platform.
The raw scripts of each pipeline and their DNAnexus application counterpart are available at this repository: https://gitlab.com/danio-code/public
All the pipelines used for processing the data use Ensembl genome GrCz10 / UCSC danRer10 for mapping.
Access to the data
All the data related to freeze-01 are accessible via the data export page of the DCC, by selecting the “freeze-01” version option in the left panel.
Please note that an user account on the DCC platform is still necessary to view and download the data. If you haven't registered yet, we invite you to sign up and contact the administrator of the DCC.
Update 2017-08-04: Data is currently not available. Only fastq sequencing files, track files and counts per tag cluster will be available to public for freeze-01
Update 2017-10-16: The data for freeze-01 will include: the fastq sequencing files, The quality control / statistical analysis results of the data, the clustering outputs, the track hub files for UCSC Genome Browser visualization. Those data are currently under preparation.
Update 2018-06-24: A user account on the DCC is not necessary anymore to download data. A user account is only necessary to upload annotations and data.

What is next...
The DANIO-CODE DCC is still a work in progress. You will notice the data available for the freeze is very unequal in terms of quantity and quality from one category to another. Amongst the different developmental stages, the number of available RNA-seq samples is unevenly distributed, including stages without any RNA-seq data.
Our goal for the next data freeze (freeze-02) will be to expand the database to fill these gaps. We are now trying to widen the range of development stages and biosample types, by looking into publicly available datasets in the literature. We also seek your help to complete our data coverage; if you would like to provide us with more data, we would be glad to be contacted by you.
Last but not least, new pipelines will be developed and implemented for freeze-02, making more assay types available to public.
If you would like to contribute to this project, need help, information or have any comments or suggestions, we invite you to contact us at the following e-mail address: michael.dong@ki.se.
We would like to thank all the DANIO-CODE consortium partners and colleagues for their support, as well as the collaborators who provided us with data for this important stage of the project.
We are grateful for all your contributions. Be sure that we will make our best efforts in carrying this project forward.
With best regards,
The DANIO-CODE DCC team
DANIO-CODE DCC technical staff:
- Damir Baranašić, pipeline developer (ATAC-seq, BS-seq and ChIP-seq)
- Michaël Dong, pipeline developer (CAGE-seq), in charge of freeze-01
- Matthias Hörtenhuber, DCC Lead Developer and Administrator
- Abdul Kadir Mukarram, DCC Co-administrator and pipeline developer (RNA-seq)
Additional Information
List of series included in freeze-01
| Series name | Series ID/link | Date of raw data upload | Assay type | Team | Laboratory/Institution, Country |
|---|---|---|---|---|---|
| ATAC-seq on 24hpf whole embryos | DCD000072SR | May 6th, 2016 | ATAC-seq | Skarmeta Lab | Centro Andaluz de Biología del Desarrollo, Spain |
| Genes enriched in pancreatic ductal and beta cells | (private) | May 6th, 2016 | RNA-seq | Peers Lab | University of Liège, Belgium |
| CHIP-seq in early development | DCD000136SR | May 6th, 2016 | ChIP-seq | Skarmeta Lab | Centro Andaluz de Biología del Desarrollo, Spain |
| Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. | DCD000139SR | May 6th, 2016 | RNA-seq | Giraldez Lab | Yale School of Medicine, USA |
| Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. | DCD000141SR | May 6th, 2016 | RNA-seq | Giraldez Lab | Yale School of Medicine, USA |
| Upstream ORFs are prevalent translational repressors in vertebrates. | DCD000142SR | May 6th, 2016 | RNA-seq | Giraldez Lab | Yale School of Medicine, USA |
| Eomesodermin targets at sphere stage with RNA-seq | DCD000145SR | May 9th, 2016 | RNA-seq | Wardle Lab | King’s College London, UK |
| Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition | DCD000170SR | May 16th, 2016 | RNA-seq | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
| Zic3 interacts with distant regulatory elements to regulate zebrafish developmental genes | DCD000171SR | May 16th, 2016 | ChIP-seq | Mathavan Lab | Nanyang Technological University, Medical School, Singapore |
| The Biotagging toolkit for analysis of specific cell populations reveals gene regulatory logic encoded in the nuclear transcriptome (under review) | DCD000177SR | May 18th, 2016 | RNA-seq | Sauka-Spengler Lab | University of Oxford, UK |
| Pioneering chromatin for neural crest specification (working title) | (private) | May 30th, 2016 | RNA-seq | Sauka-Spengler Lab | University of Oxford, UK |
| Genome-wide maps of binding sites of Nanog-like and Mxtx2 in blastula stage zebrafish embryos | DCD000204SR | June 2nd, 2016 | ChIP-seq | Zon Lab | Boston Children Hospital, USA |
| A Cdx4-Sall4 regulatory module controls the transition from mesoderm formation to embryonic hematopoiesis | DCD000205SR | June 2nd, 2016 | ChIP-seq | Zon Lab | Boston Children Hospital, USA |
| Zebrafish Globin Locus (ChIP-seq) | DCD000206SR | June 2nd, 2016 | ChIP-seq | Zon Lab | Boston Children Hospital, USA |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [zebrafish ChIP-seq] | DCD000207SR | June 2nd, 2016 | ChIP-seq | Zon Lab | Boston Children Hospital, USA |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [zebrafish RNA-Seq] | DCD000208SR | June 2nd, 2016 | RNA-seq | Zon Lab | Boston Children Hospital, USA |
| A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation [ATAC-Seq] | DCD000209SR | June 2nd, 2016 | ATAC-seq | Zon Lab | Boston Children Hospital, USA |
| Comprehensive identification of long non-coding RNAs expressed during zebrafish embryogenesis | DCD000225SR | June 9th, 2016 | RNA-seq | Schier Lab | Harvard University, USA |
| DNA methylation reprogramming during early zebrafish embryo development | DCD000227SR | June 11th, 2016 | RNA-seq | Liu Lab | Beijing Institute of Genomics, CAS, China |
| Comprehensive maps of DNA methylation in mature gametes and at various embryonic stages of cleavage phase zebrafish development. | DCD000229SR | June 16th, 2016 | BS-seq | Cairns Lab | Howard Hughes Medical Institute, USA |
| Head/trunk CAGE | (private) | July 27th, 2016 | CAGE-seq | Mueller lab | University of Birmingham, UK |
| H2A.Z coverage for 30% epiboly | (private) | July 27th, 2016 | ChIP-seq | Mueller lab | University of Birmingham, UK |
| CAGE data for developmental time course | DCD000242SR | July 27th, 2016 | CAGE-seq | Mueller lab | University of Birmingham, UK |
| Embryonic promoterome | DCD000243SR | July 27th, 2016 | RNA-seq | Mueller lab | University of Birmingham, UK |
| Analysis of open chromatin in early development | (private) | July 27th, 2016 | ATAC-seq | Mueller lab | University of Birmingham, UK |
| Loss of function of myosin chaperones triggers Hsf1-mediated transcriptional response in skeletal muscle cells | DCD000247SR | Sept. 19th, 2016 | RNA-seq | Strähle Lab | Karlsruhe Institute of Technology, Germany |
| Baseline expression from transcriptional profiling of zebrafish developmental stages | DCD000324SR | May 2nd, 2017 | RNA-seq | Busch-Nentwich Lab | Wellcome Trust Sanger Institute, UK |
List of data providers for freeze-01
- Busch-Nentwich Lab, Wellcome Trust Sanger Institute, UK
- Cairns Lab, Howard Hughes Medical Institute, USA
- Giraldez Lab, Yale School of Medicine, USA
- Liu Lab, Beijing Institute of Genomics, CAS, China
- Mathavan Lab, Nangyang Technological University, Medical School, Singapore
- Mueller Lab, University of Birmingham, UK
- Peers Lab, University of Liège, Belgium
- Sauka-Spengler Lab, University of Oxford, UK
- Schier Lab, Harvard University, USA
- Skarmeta Lab, Centro Andaluz de Biologia del Desarrollo, Spain
- Strähle Lab, Karlsruhe Institute of Technology, Germany
- Wardle Lab, King’s College London, UK
- Zon Lab, Boston Children Hospital, USA